转录组和蛋白组如何关联分析?先从绘制九象限图开始
五种常用蛋白质组学定量分析方法对比 - 知乎 (zhihu.com)
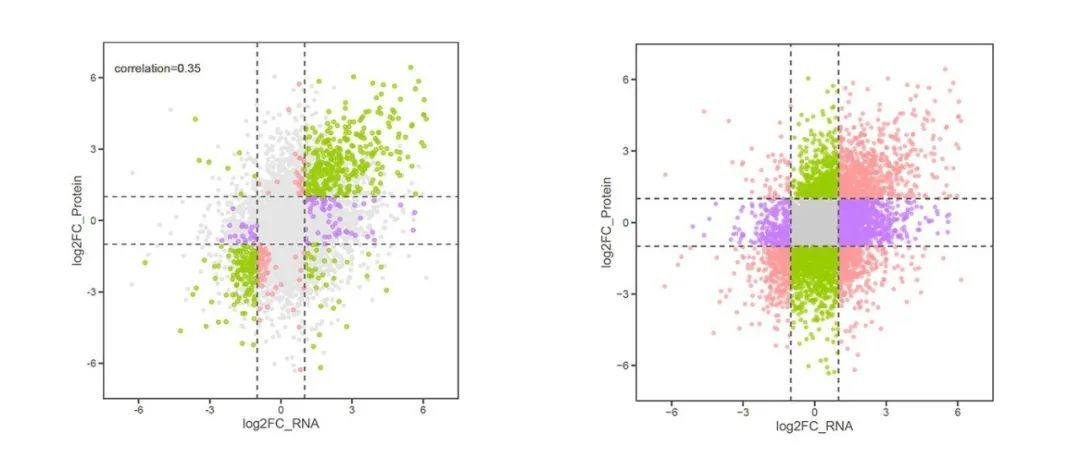
九象限图在多组学关联分析中非常重要,例如我们可以用九象限图展示“转录组+蛋白组”、“转录组+翻译组”等关联分析中不同基因的差异表达情况。通过分析两种组学数据的相关性,我们可进一步缩小候选基因的范围。

当然,之前的微信文章《Graphpad Prism能不能画九象限图?》中介绍过九象限图的画法,下面再为大家介绍一种效果更好看的画法。
1. 数据合并
#读入两个差异分析表(分别是转录组和翻译组)并合并;
#设置工作目录;
setwd("C:/Users/86136/Desktop/九象限图的绘制")
#查看工作目录中的文件;
dir()
#[1] "protein_exp.csv" "RNA_exp.csv"
#读入数据;
RNA =read.csv("RNA_exp3.csv",header=T)
protein=read.csv("protein_exp3.csv",header=T)#查看数据的维度;
dim(RNA)
#[1] 10565 5
#查看前6行;
head(RNA)
dim(protein)
#[1] 7028 5
#查看前6行;
head(protein)
#合并两个表格。选择表头为"id"的列进行合并(取交集);
#suffixes:如果两个表列名相同,则会在列名后加入suffixes(后缀)参数中对应的后缀;
#all.x=FALSE,all.y=FALSE,表示输出的是x,y表格的交集;
combine= merge(RNA,protein,
by.x="id",
by.y="id",
suffixes = c("_RNA","_Protein") ,
all.x=FALSE,
all.y=FALSE)
#查看合并后表格的维度;
dim(combine)
#[1] 6657 9
#保存合并后的数据;
write.csv(combine,"RNA_protein3.csv",row.names=FALSE)2. 数据筛选
#载入相关的R包;
library(dplyr)
library(ggplot2)
library(ggrepel)#提取用于作图的列组成新数据框;
data <- data.frame(combine[c(1,3,4,6,7,8)])
#查看前6行;
head(data)
#对数据进行分组;
#生成显著上下调数据标签;
data$part <- case_when(abs(data$log2FC_RNA) >= 1 & abs(data$log2FC_Protein) >= 1 ~ "part1379",
abs(data$log2FC_RNA) < 1 & abs(data$log2FC_Protein) > 1 ~ "part28",
abs(data$log2FC_RNA) > 1 & abs(data$log2FC_Protein) < 1 ~ "part46",
abs(data$log2FC_RNA) < 1 & abs(data$log2FC_Protein) < 1 ~ "part5")
head(data)
3. 绘制表格
#开始尝试绘图;
p0 <-ggplot(data,aes(log2FC_RNA,log2FC_Protein,color=part))
#添加散点;
p1 <- p0+geom_point(size=1.2)+guides(color="none")
p1
#自定义半透明颜色;
mycolor <- c("#FF9999","#99CC00","#c77cff","gray80")
p2 <- p1 + scale_colour_manual(name="",values=alpha(mycolor,0.7))
p2
#添加辅助线;
p3 <- p2+geom_hline(yintercept = c(-1,1),
size = 0.5,
color = "grey40",
lty = "dashed")+
geom_vline(xintercept = c(-1,1),
size = 0.5,
color = "grey40",
lty = "dashed")
p3
#调整横轴和纵轴绘图区域的范围;
#设置y轴范围(上下两端的空白区域设为1),修改刻度标签;
#expansion函数设置坐标轴范围两端空白区域的大小;mult为“倍数”模式,add为“加性”模式;
p4<-p3+
scale_y_continuous(expand=expansion(add = c(0.5, 0.5)),
limits = c(-7, 7),
breaks = c(-6,-3,0,3,6),
label = c("-6","-3","0","3","6"))+
scale_x_continuous(expand=expansion(add = c(0.5, 0.5)),
limits = c(-7, 7),
breaks = c(-6,-3,0,3,6),
label = c("-6","-3","0","3","6"))
p4
#自定义图表主题,对图表做精细调整;
top.mar=0.2
right.mar=0.2
bottom.mar=0.2
left.mar=0.2
#隐藏纵轴,并对字体样式、坐标轴的粗细、颜色、刻度长度进行限定;
mytheme<-theme_bw()+
theme(text=element_text(family = "sans",colour ="gray30",size = 12),
panel.grid = element_blank(),
panel.border = element_rect(size = 0.8,colour = "gray30"),
axis.line = element_blank(),
axis.ticks = element_line(size = 0.6,colour = "gray30"),
axis.ticks.length = unit(1.5,units = "mm"),
plot.margin=unit(x=c(top.mar,right.mar,bottom.mar,left.mar),
units="inches"))
#应用自定义主题;
p5 <- p4+mytheme
p5
#如果考虑差异的显著性,则需要进一步分组;
#生成至少在一个组学显著上下调的数据标签;
data$sig <- case_when(data$Pvalue_RNA < 0.05 & data$Pvalue_Protein <0.05 ~ "sig",
data$Pvalue_RNA >= 0.05 | data$Pvalue_Protein >=0.05 ~ "no")
head(data)
#将作图数据表格拆分成显著和不显著两部分;
sig <- filter(data,sig == "sig")
non <- filter(data,sig == "no")#重新进行绘图;
p6 <-ggplot(data,aes(log2FC_RNA,log2FC_Protein))+
geom_point(data=non,aes(log2FC_RNA,log2FC_Protein),size=1.2,color="gray90")+
geom_point(data=sig,aes(log2FC_RNA,log2FC_Protein,color=part),size=1.5)+
guides(color="none")
p6
#自定义半透明颜色;
mycolor <- c("#99CC00","#FF9999","#c77cff","gray90")
p7 <- p6 + scale_colour_manual(name="",values=alpha(mycolor,0.7))+mytheme
p7
#添加辅助线并修改坐标轴范围;
p8 <- p7+geom_hline(yintercept = c(-1,1),
size = 0.5,
color = "gray40",
lty = "dashed")+
geom_vline(xintercept = c(-1,1),
size = 0.5,
color = "gray40",
lty = "dashed")+
scale_y_continuous(expand=expansion(add = c(0.5, 0.5)),
limits = c(-7, 7),
breaks = c(-6,-3,0,3,6),
label = c("-6","-3","0","3","6"))+
scale_x_continuous(expand=expansion(add = c(0.5, 0.5)),
limits = c(-7, 7),
breaks = c(-6,-3,0,3,6),
label = c("-6","-3","0","3","6"))
p8
#计算两个组学差异倍数的相关性,并取2位小数;
cor = round(cor(data$log2FC_RNA,data$log2FC_Protein),2)
#准备作为图形的标题;
lab = paste("correlation=",cor,sep="")
lab
#[1] "correlation=0.35"
#在图上添加文字标签;
p8+geom_text(x = -5.2, y = 6.4, label = lab, color="gray40")
好啦,今天先分享到这里啦~
想要第一时间获取科研绘图干货,欢迎关注基迪奥生物旗下的官方工作号:SCIPainter
专注分享科研绘图技能与工具!






