自己也是个小白,初次安装org.Hs.eg.db碰到好多问题,步骤总结如下:
第一,instal.packages()安装,不行则换 BiocManager::install()
第二,如BiocManager不成,说明不是最新版本,可按提示安装最新版
BiocManager::install(version = '3.13') #由于R版本较新,按照提示中的相关信息,安装最新版本的第三,再次安装
BiocManager::install("org.Hs.eg.db")第四,之后每个人碰到问题就不一样了,我遇到的问题是
类似于包不能因移除,而无法更新(有11个警告信息,不详细列举)
Bioconductor version 3.13 (BiocManager 1.30.16), R 4.1.0 (2021-05-18)
Old packages: 'gert', 'RCurl', 'readr', 'survival', 'XML'
Update all/some/none? [a/s/n]:
a
#更新过后报错(中间代码省略)
package ‘stringi’ successfully unpacked and MD5 sums checked
Warning: cannot remove prior installation of package ‘stringi’
Warning: restored ‘stringi’解决办法:packages——找到与报警信息对应包——点击❌去除相应包(之后可再安装)
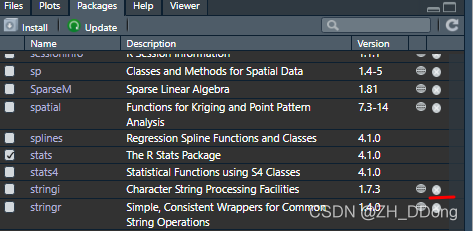
第五,再次BiocManager::install("org.Hs.eg.db")
第六,最后library("org.Hs.eg.db"),出现以下代码,恭喜你安装成功
载入需要的程辑包:AnnotationDbi
载入需要的程辑包:stats4
载入需要的程辑包:BiocGenerics
载入需要的程辑包:parallel载入程辑包:‘BiocGenerics’The following objects are masked from ‘package:parallel’:clusterApply, clusterApplyLB, clusterCall, clusterEvalQ,clusterExport, clusterMap, parApply, parCapply, parLapply,parLapplyLB, parRapply, parSapply, parSapplyLBThe following objects are masked from ‘package:stats’:IQR, mad, sd, var, xtabsThe following objects are masked from ‘package:base’:anyDuplicated, append, as.data.frame, basename, cbind, colnames,dirname, do.call, duplicated, eval, evalq, Filter, Find, get,grep, grepl, intersect, is.unsorted, lapply, Map, mapply, match,mget, order, paste, pmax, pmax.int, pmin, pmin.int, Position,rank, rbind, Reduce, rownames, sapply, setdiff, sort, table,tapply, union, unique, unsplit, which.max, which.min载入需要的程辑包:Biobase
Welcome to BioconductorVignettes contain introductory material; view with'browseVignettes()'. To cite Bioconductor, see'citation("Biobase")', and for packages 'citation("pkgname")'.载入需要的程辑包:IRanges
载入需要的程辑包:S4Vectors载入程辑包:‘S4Vectors’The following objects are masked from ‘package:base’:expand.grid, I, unname载入程辑包:‘IRanges’The following object is masked from ‘package:grDevices’:windows