数据和代码获取:请查看主页个人信息!!!
大家好,今天我将介绍高分杂志组合图绘制,图形来源于Nature的一篇文章:

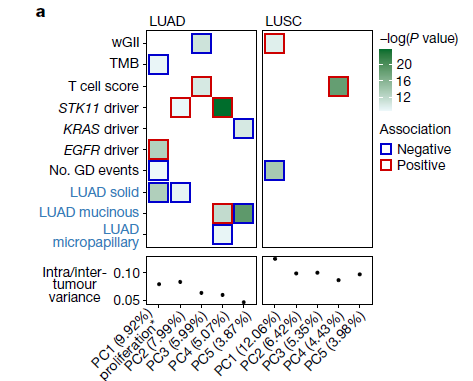
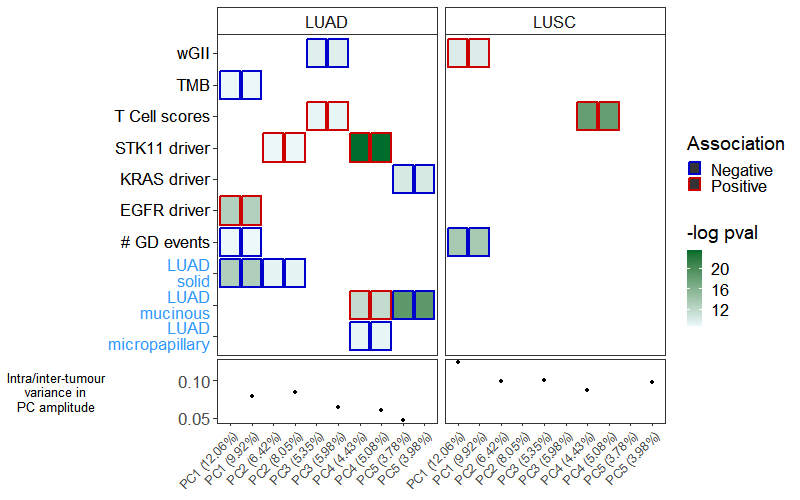
该图展示了转录组多样性的主成分(PCs)与基因组(黑色标签)和临床(蓝色标签)变量之间的关系。原文使用混合效应线性模型进行显著性的确定,感兴趣的同学可以进行深入学习。
本文主要对可视化方法进行展示,话不多说,我们直接开始:
Step1:数据载入
rm(list=ls())pacman::p_load(tidyverse,aplot,ggtext)data <- read.csv('data.csv', row.names = 1)
大家可以看到:原文图中有三组Y文本非黑色字体,这个并非P图实现,而是通过【ggtext包】实现,因此我们在绘图前需要对数据分组颜色进行赋值。

Step2:定义Y轴字体颜色
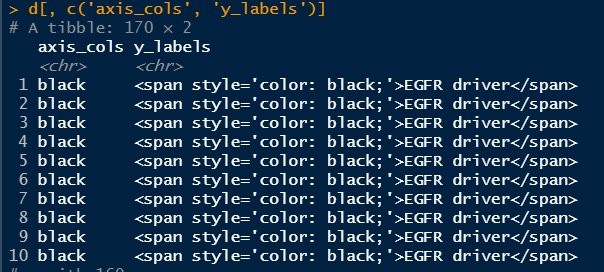
d <-data %>%filter(variable_type %in% c("genomic", "clinical")) %>%mutate(axis_cols = if_else(variable_type == "clinical",true = "#3399FF",false = "black")) %>%mutate(y_labels = paste0("<span style='color: ",axis_cols,";'>",variable,"</span>")) %>%group_by(variable) %>%filter(!all(is.na(get('log_pval'))))d[, c('axis_cols', 'y_labels')]
"<span style='color: ", axis_cols, ";'>", variable, "</span>" 是一段用于创建一个带有指定颜色的标签。axis_cols 是指颜色,variable 表示要显示的标签文本。通过将这两个变量与一些HTML标签和样式属性组合在一起,可以生成一个具有指定颜色的标签。在这个特定的代码段中,使用了 <span> 标签和 color 样式属性来设置标签的颜色。
Step3:绘制主热图
p1 <- ggplot(d, aes(x = pc_label, y = y_labels, fill = log_pval)) +geom_tile(aes(color = effect_direction, width = 0.9, height = 0.9), size = 1) +facet_grid(. ~ histology, scales = "free") +scale_fill_gradient(low = "#edf8fb", high = "#006d2c", na.value = "white",name = '-log pval') +scale_color_manual(values = c("blue3", "red3"), name = "Association",na.value = "white", na.translate = FALSE) +theme_bw() +theme(axis.text.x = element_blank(),axis.title.x = element_blank(),axis.ticks.x = element_blank(),axis.text.y = element_markdown(size = 13),axis.title.y = element_blank(),legend.title = element_text(size = 14),legend.text = element_text(size = 13),panel.grid.major = element_blank(),panel.grid.minor = element_blank(),strip.background = element_rect(fill = "white"),strip.text = element_text(size = 13),legend.position = 'right',legend.key.size= unit(0.4, 'cm'))p1
上面代码中【ggtext包的element_markdown】函数,实现了Y轴字体颜色的映射,这是一个很好的绘图技巧,能增加图形的美观程度。

Step4:绘制子图
p2 <- ggplot(d, aes(x = pc_label, y = median_ratio_var)) +geom_point(size = 1) +theme_bw() +scale_y_continuous(breaks = scales::pretty_breaks(n = 3)) +facet_grid(. ~ Histology, scales = "free") +ylab("Intra/inter-tumour\nvariance in\nPC amplitude") +theme(axis.text.x=element_text(angle=45,vjust=1, hjust=1),axis.title.x = element_blank(),axis.text.y = element_text(size = 13),axis.title.y = element_text(size = 10, angle=0, vjust=0.5),legend.title = element_text(size = 14),legend.text = element_text(size = 13),strip.background = element_blank(),strip.text = element_blank(),panel.grid.major = element_blank(),panel.grid.minor = element_blank(),plot.margin = unit(c(0, 0, 0, 0), "null"))p2
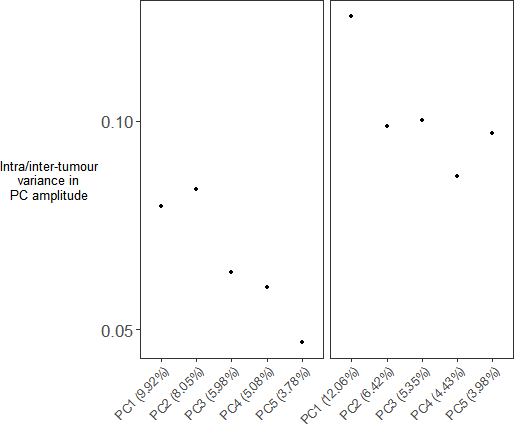
Step5:拼图
p1 %>%insert_bottom(p2, height = 0.2)ggsave('pic1.pdf', width = 9, height = 7)
当然,我们也可以在原图的基础上,进一步展示更多数据内容:
Step6:个性化延伸
p3 <-ggplot(d, aes(x = y_labels, y = r_marg, color = r_marg, fill = r_marg), size = abs(r_marg)) +geom_boxplot(width = 0.35, color = 'grey50') +geom_point(size = 1) +coord_flip() +ggsci::scale_fill_material("blue") +ggsci::scale_color_material("blue") +xlab('') +labs(fill = 'Correlation', color = 'Correlation', title = 'Correlation') +scale_y_continuous(breaks = scales::pretty_breaks(n = 3)) +theme_bw() +theme(axis.text.x=element_text(angle=45,vjust=1, hjust=1),axis.title.x = element_blank(),axis.ticks.y = element_blank(),axis.text.y = element_blank(),axis.title.y = element_text(size = 10, angle=0, vjust=0.5),legend.title = element_text(size = 14),legend.text = element_text(size = 13),strip.background = element_blank(),strip.text = element_blank(),panel.grid.major = element_blank(),panel.grid.minor = element_blank(),plot.margin = unit(c(0, 0, 0, 0), "null"),legend.key.size= unit(0.4, 'cm'))p3p1 %>%insert_bottom(p2, height = 0.2) %>%insert_right(p3, width = 0.2)ggsave('pic2.pdf', width = 10, height = 7)
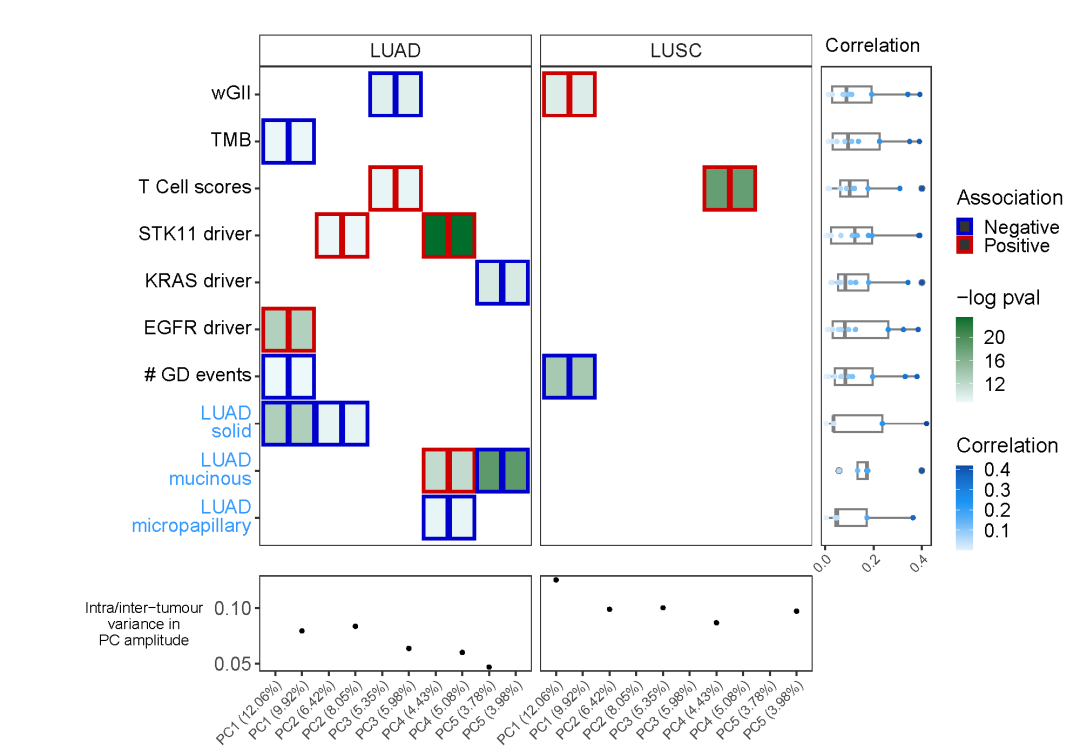
好啦,本期分享就到这里啦!喜欢本期内容的童鞋们后台回复“相关性组合图”获得本期代码和数据。