library(survival)
library(randomForestSRC)
help(package="randomForestSRC")
#构建普通的随机生存森林
data(cancer,package="survival")
lung$status<-lung$status-1
rfsrc.fit1 <- rfsrc(Surv(time, status) ~ ., lung,ntree = 100,block.size = 1,seed=123)
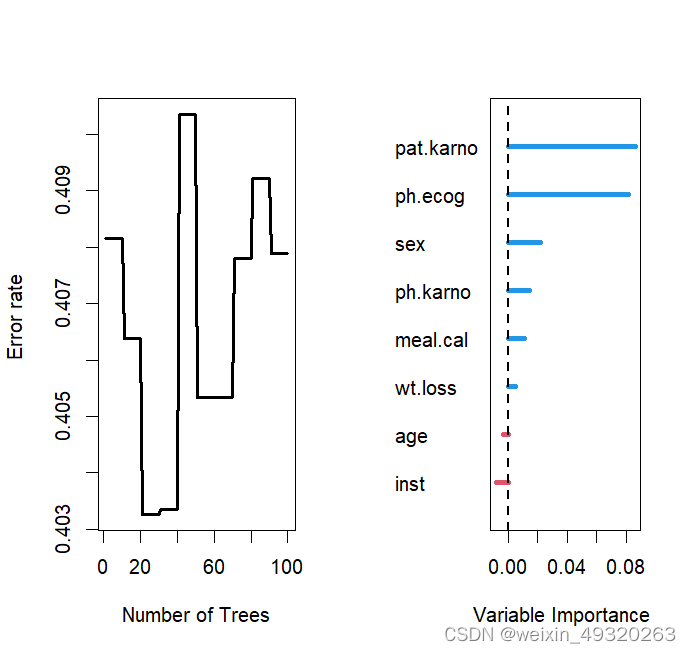
plot.rfsrc(rfsrc.fit1)+title("误差曲线")#绘制误差曲线
plot.survival(rfsrc.fit1)+title("生存估计")#绘制生存估计:1、每个个体的生存估计2、Brier评分3、连续秩概率分数(CRPS)=Brier分数/时间。4、个体死亡率与观察时间关系图
plot.survival.rfsrc(rfsrc.fit1)
1-rfsrc.fit1$err.rate[rfsrc.fit1$ntree]#C指数0.5761229
#与Cox回归对比C指数
options("na.action")
lung<-na.omit(lung)
cox1<-coxph(Surv(time, status)~.,lung)
1-get.cindex(lung$time,lung$status,predict(cox1,lung))#0.6482742


#构建随机生存森林模型-竞争风险
data(wihs, package = "randomForestSRC")
table(wihs$status)
rfsrc.fit2 <- rfsrc(Surv(time, status) ~ ., wihs,ntree = 100)
plot.competing.risk(rfsrc.fit2)
1-rfsrc.fit2$err.rate[rfsrc.fit2$ntree]#0.6079373
#进行预测新数据/生成生存率预测
pred <- predict(rfsrc.fit2, newdata = wihs, OOB = TRUE, prediction = TRUE, importance = TRUE, proximity = TRUE, maxnodes = 10)

#绘制校准曲线
library(riskRegression)
rf_fit<-riskRegression::Score(list("fit1" = rfsrc.fit1,"fit2"=cox1),formula = Surv(time, status) ~ 1,data = lung, # 测试集plots = "calibration",conf.int = T,B = 500, #重抽样500次 #交叉验证M = 40,#抽样样本量 #交叉验证times=c(100) # 时间)
args(plotCalibration)
riskRegression::plotCalibration(rf_fit,cens.method="local",xlab = "Predicted Risk",ylab = "Observerd RISK",col=c("red","blue"),legend=T)

#实现随机生存森林参数调优
tune(Surv(time, status) ~ ., lung,seed=123)#最佳nodesize4 mtry2
#筛选重要预测因素
var<-var.select(object=rfsrc.fit1,method="md",#变量筛选方法conservative="low"#筛选阈值)
top<-var$topvars
#变量重要性
vimp(rfsrc.fit1) %>% plot








![[HDLBits] Exams/m2014 q4c](https://img-blog.csdnimg.cn/img_convert/5bccfb0c6b59e6a4ea6bc98f71998fe9.png)